代謝
Metabolism/ja


| Part of a series on |
| Biochemistry/ja |
|---|
 |
代謝(/məˈtæbəlɪzəm/, from Greek: μεταβολή metabolē, "change")とは、生物における生命を維持するための一連の化学反応のことである。代謝の3つの主な機能は、食物中のエネルギーを細胞プロセスを実行するために利用可能なエネルギーに変換すること、食物をタンパク質、脂質、核酸、および一部の炭水化物の構成要素に変換すること、および代謝廃棄物を除去することである。 これらの酵素触媒反応により、生物は成長・繁殖し、構造を維持し、環境に対応することができる、 また、代謝という言葉は、消化を含む生物で起こる全ての化学反応の総体を指すこともある、 この場合、上記の細胞内の一連の反応は中間(または中間代謝)と呼ばれる。
代謝反応は、異化-化合物の分解(例えば、細胞呼吸によるグルコースからピルビン酸への分解)と、同化-化合物(タンパク質、炭水化物、脂質、核酸など)の構築(合成)に分類される。通常、異化はエネルギーを放出し、同化はエネルギーを消費する。
代謝の化学反応は代謝経路に組織化されており、そこではある化学物質が一連のステップを経て別の化学物質に変換され、各ステップは特定の酵素によって促進される。酵素は、エネルギーを放出する自発的反応に結合させることによって、エネルギーを必要とし、それ自体では起こらないような望ましい反応を生物が推進することを可能にするので、代謝にとって極めて重要である。酵素は触媒として機能し、反応をより速く進行させる。また、例えばセルの環境の変化や他の細胞からのシグナルに応答して、代謝反応の速度を調節することもできる。
特定の生物の代謝システムは、どの物質を栄養とし、どの物質を毒とするかを決定する。例えば、いくつかの原核生物は硫化水素を栄養としているが、このガスは動物にとっては毒である。生物の基礎代謝量は、これらすべての化学反応によって消費されるエネルギー量の尺度である。
代謝の顕著な特徴は、大きく異なる種の間で基本的な代謝経路が類似していることである。例えば、クエン酸サイクルの中間体として最もよく知られているカルボン酸のセットは、すべての既知の生物に存在し、単細胞細菌の大腸菌やゾウのような巨大な多細胞生物などの多様な種に見られる。このような代謝経路の類似性は、進化史において早くから出現していたためと考えられ、その保持はその効能によるものと考えられる。2型糖尿病、メタボリックシンドローム、がんなどの様々な疾患では、正常な代謝が破綻している。がん細胞の代謝もまた正常細胞の代謝とは異なっており、こうした違いを利用してがんの治療介入のターゲットを見つけることができる。
主要な性化学物質


動物、植物、微生物を構成する構造のほとんどは、4つの基本的なクラスの分子からできている: アミノ酸、炭水化物、核酸、脂質(しばしば脂肪と呼ばれる)である。これらの分子は生命維持に不可欠であるため、代謝反応は細胞や組織を作る際にこれらの分子を作ることに集中するか、消化によって分解してエネルギーを得るために使用することに集中する。これらの生化学物質は結合して、DNAやタンパク質といった生命に必須な高分子のポリマーを作ることができる。
| 分子の種類 | モノマーの形態名 | ポリマーの形態名 | ポリマー形態の例 |
|---|---|---|---|
| アミノ酸 | アミノ酸 | プロテイン (ポリペプチドでできている) | 繊維状タンパク質 と 球状タンパク質 |
| 炭水化物 | 単糖類 | 多糖類 | スターチ, グリコーゲン, セルロース |
| 核酸 | ヌクレオチド | ポリヌクレオチド | DNA と RNA |
アミノ酸とタンパク質
タンパク質はアミノ酸がペプチド結合で結合した直鎖状に配列したものである。多くのタンパク質は酵素であり、代謝における化学反応を触媒する。 他のタンパク質は、細胞の形状を維持する足場のシステムである細胞骨格を形成するものなど、構造的または機械的な機能を持つ。 タンパク質はまた、細胞シグナル伝達、免疫応答、細胞接着、膜を介した活性輸送、および細胞周期において重要である。 アミノ酸はまた、クエン酸サイクル(トリカルボン酸サイクル)に入るための炭素源を提供することで、細胞のエネルギー代謝にも寄与する、 特に、グルコースのような主要なエネルギー源が不足しているときや、細胞が代謝ストレスを受けているときには、この役割を果たす。
脂質
脂質は生化学物質の中で最も多様なグループである。その主な構造的用途は、細胞膜のような内外の生体膜の一部としての利用である。その化学エネルギーを利用することもできる。脂質は、酸素を含む小さな極性領域を持つ長い非極性炭化水素鎖を含む脂肪酸のポリマーである。脂質は通常疎水性または両親媒性の生体分子と定義されるが、エタノール、ベンゼン、クロロホルムなどの有機溶媒に溶解する。脂肪は脂肪酸とグリセロールを含む化合物の大きなグループであり、エステル結合によって3つの脂肪酸に結合したグリセロール分子はトリアシルグリセリドと呼ばれる。この基本構造にはいくつかのバリエーションがあり、スフィンゴミエリンのsphingosine/jaスフィンゴシンのような骨格や、リン脂質のようなリン酸のような親水性|親水性基がある。ステロールなどのステロイドも脂質の主要な分類である。
炭水化物
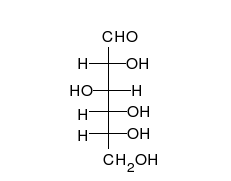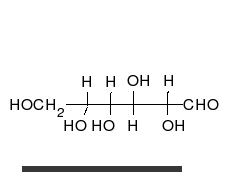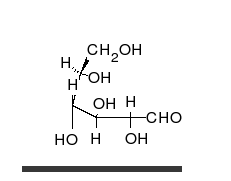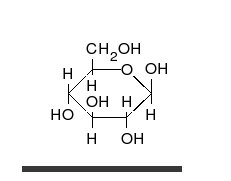
炭水化物は、多くの水酸基が結合したアルデヒドまたはケトンであり、直鎖または環として存在することができる。炭水化物は最も豊富な生体分子であり、エネルギーの貯蔵や輸送(デンプン、グリコーゲン)、構造成分(植物のセルロース、動物のキチン)など、多くの役割を担っている。基本的な炭水化物単位は単糖と呼ばれ、ガラクトース、フルクトース、そして最も重要なグルコースが含まれる。単糖類は連結してほとんど無限の方法で多糖類を形成することができる。
核酸
DNAとRNAという2つの核酸は、核酸のポリマーである。各核酸は、リボースまたはデオキシリボースの糖基に結合したリン酸から構成され、窒素塩基に結合している。核酸は、遺伝情報の保存と利用、および転写とタンパク質生合成の過程を通してのその解釈に重要である。この情報はDNA修復機構によって保護され、DNA複製によって伝播する。多くのウイルスはRNAゲノムを持っており、例えばHIVは逆転写を用いてそのウイルスRNAゲノムからDNAテンプレートを作り出す。スプライソソームやリボソームなどのリボザイムに含まれるRNAは、化学反応を触媒できるので酵素に似ている。個々の核酸はリボース糖に核酸塩基を結合させて作られる。これらの塩基は窒素を含む複素環であり、プリンまたはピリミジンに分類される。ヌクレオチドはまた、代謝基転移反応における補酵素としても働く。
補酵素(コエンザイム)

代謝には膨大な数の化学反応が含まれるが、そのほとんどは、分子内の原子の官能基とその結合の移動を伴う、いくつかの基本的なタイプの反応に分類される。この一般的な化学反応によって、細胞は小さな代謝中間体のセットを使って、異なる反応間で化学基を運ぶことができる。これらの基転移中間体は補酵素と呼ばれる。各クラスの基転移反応は、特定の補酵素によって行われる。補酵素は、それを生成する一連の酵素の基質であり、それを消費する一連の酵素の基質でもある。 したがって、これらの補酵素は絶えず作られ、消費され、そして再利用される。
中心的な補酵素のひとつが、細胞のエネルギー通貨であるアデノシン三リン酸(ATP)である。このヌクレオチドは、異なる化学反応間で化学エネルギーを伝達するために使われる。細胞内に存在するATPの量はわずかだが、絶えず再生されるため、人体は1日に自分の体重ほどのATPを使うことができる。ATPは異化作用と同化作用の橋渡しをする。異化作用によって分子が分解され、同化作用によって分子が組み合わされる。異化反応はATPを生成し、同化反応はATPを消費する。また、リン酸化反応ではリン酸基の運搬役としても働く。
A vitamin is an organic compound needed in small quantities that cannot be made in cells. In human nutrition, most vitamins function as coenzymes after modification; for example, all water-soluble vitamins are phosphorylated or are coupled to nucleotides when they are used in cells. Nicotinamide adenine dinucleotide (NAD+), a derivative of vitamin B3 (niacin), is an important coenzyme that acts as a hydrogen acceptor. Hundreds of separate types of dehydrogenases remove electrons from their substrates and reduce NAD+ into NADH. This reduced form of the coenzyme is then a substrate for any of the reductases in the cell that need to transfer hydrogen atoms to their substrates. Nicotinamide adenine dinucleotide exists in two related forms in the cell, NADH and NADPH. The NAD+/NADH form is more important in catabolic reactions, while NADP+/NADPH is used in anabolic reactions.

Mineral and cofactors
Inorganic elements play critical roles in metabolism; some are abundant (e.g. sodium and potassium) while others function at minute concentrations. About 99% of a human's body weight is made up of the elements carbon, nitrogen, calcium, sodium, chlorine, potassium, hydrogen, phosphorus, oxygen and sulfur. Organic compounds (proteins, lipids and carbohydrates) contain the majority of the carbon and nitrogen; most of the oxygen and hydrogen is present as water.
The abundant inorganic elements act as electrolytes. The most important ions are sodium, potassium, calcium, magnesium, chloride, phosphate and the organic ion bicarbonate. The maintenance of precise ion gradients across cell membranes maintains osmotic pressure and pH. Ions are also critical for nerve and muscle function, as action potentials in these tissues are produced by the exchange of electrolytes between the extracellular fluid and the cell's fluid, the cytosol. Electrolytes enter and leave cells through proteins in the cell membrane called ion channels. For example, muscle contraction depends upon the movement of calcium, sodium and potassium through ion channels in the cell membrane and T-tubules.
Transition metals are usually present as trace elements in organisms, with zinc and iron being most abundant of those. Metal cofactors are bound tightly to specific sites in proteins; although enzyme cofactors can be modified during catalysis, they always return to their original state by the end of the reaction catalyzed. Metal micronutrients are taken up into organisms by specific transporters and bind to storage proteins such as ferritin or metallothionein when not in use.
Catabolism
Catabolism is the set of metabolic processes that break down large molecules. These include breaking down and oxidizing food molecules. The purpose of the catabolic reactions is to provide the energy and components needed by anabolic reactions which build molecules. The exact nature of these catabolic reactions differ from organism to organism, and organisms can be classified based on their sources of energy, hydrogen, and carbon (their primary nutritional groups), as shown in the table below. Organic molecules are used as a source of hydrogen atoms or electrons by organotrophs, while lithotrophs use inorganic substrates. Whereas phototrophs convert sunlight to chemical energy, chemotrophs depend on redox reactions that involve the transfer of electrons from reduced donor molecules such as organic molecules, hydrogen, hydrogen sulfide or ferrous ions to oxygen, nitrate or sulfate. In animals, these reactions involve complex organic molecules that are broken down to simpler molecules, such as carbon dioxide and water. Photosynthetic organisms, such as plants and cyanobacteria, use similar electron-transfer reactions to store energy absorbed from sunlight.
| Energy source | sunlight | photo- | -troph | ||
| molecules | chemo- | ||||
| Hydrogen or electron donor | organic compound | organo- | |||
| inorganic compound | litho- | ||||
| Carbon source | organic compound | hetero- | |||
| inorganic compound | auto- | ||||
The most common set of catabolic reactions in animals can be separated into three main stages. In the first stage, large organic molecules, such as proteins, polysaccharides or lipids, are digested into their smaller components outside cells. Next, these smaller molecules are taken up by cells and converted to smaller molecules, usually acetyl coenzyme A (acetyl-CoA), which releases some energy. Finally, the acetyl group on acetyl-CoA is oxidized to water and carbon dioxide in the citric acid cycle and electron transport chain, releasing more energy while reducing the coenzyme nicotinamide adenine dinucleotide (NAD+) into NADH.
Digestion
Macromolecules cannot be directly processed by cells. Macromolecules must be broken into smaller units before they can be used in cell metabolism. Different classes of enzymes are used to digest these polymers. These digestive enzymes include proteases that digest proteins into amino acids, as well as glycoside hydrolases that digest polysaccharides into simple sugars known as monosaccharides.
Microbes simply secrete digestive enzymes into their surroundings, while animals only secrete these enzymes from specialized cells in their guts, including the stomach and pancreas, and in salivary glands. The amino acids or sugars released by these extracellular enzymes are then pumped into cells by active transport proteins.

Energy from organic compounds
Carbohydrate catabolism is the breakdown of carbohydrates into smaller units. Carbohydrates are usually taken into cells after they have been digested into monosaccharides. Once inside, the major route of breakdown is glycolysis, where sugars such as glucose and fructose are converted into pyruvate and some ATP is generated. Pyruvate is an intermediate in several metabolic pathways, but the majority is converted to acetyl-CoA through aerobic (with oxygen) glycolysis and fed into the citric acid cycle. Although some more ATP is generated in the citric acid cycle, the most important product is NADH, which is made from NAD+ as the acetyl-CoA is oxidized. This oxidation releases carbon dioxide as a waste product. In anaerobic conditions, glycolysis produces lactate, through the enzyme lactate dehydrogenase re-oxidizing NADH to NAD+ for re-use in glycolysis.
Fats are catabolized by hydrolysis to free fatty acids and glycerol. The glycerol enters glycolysis and the fatty acids are broken down by beta oxidation to release acetyl-CoA, which then is fed into the citric acid cycle. Fatty acids release more energy upon oxidation than carbohydrates. Steroids are also broken down by some bacteria in a process similar to beta oxidation, and this breakdown process involves the release of significant amounts of acetyl-CoA, propionyl-CoA, and pyruvate, which can all be used by the cell for energy. M. tuberculosis can also grow on the lipid cholesterol as a sole source of carbon, and genes involved in the cholesterol-use pathway(s) have been validated as important during various stages of the infection lifecycle of M. tuberculosis.
Amino acids are either used to synthesize proteins and other biomolecules, or oxidized to urea and carbon dioxide to produce energy. The oxidation pathway starts with the removal of the amino group by a transaminase. The amino group is fed into the urea cycle, leaving a deaminated carbon skeleton in the form of a keto acid. Several of these keto acids are intermediates in the citric acid cycle, for example α-ketoglutarate formed by deamination of glutamate. The glucogenic amino acids can also be converted into glucose, through gluconeogenesis (discussed below).
Energy transformations
Oxidative phosphorylation
In oxidative phosphorylation, the electrons removed from organic molecules in areas such as the citric acid cycle are transferred to oxygen and the energy released is used to make ATP. This is done in eukaryotes by a series of proteins in the membranes of mitochondria called the electron transport chain. In prokaryotes, these proteins are found in the cell's inner membrane. These proteins use the energy from reduced molecules like NADH to pump protons across a membrane.
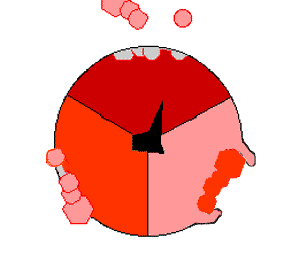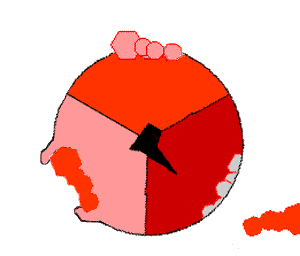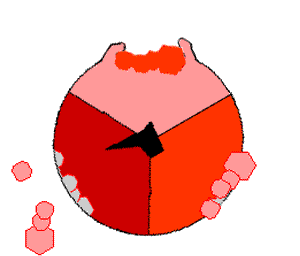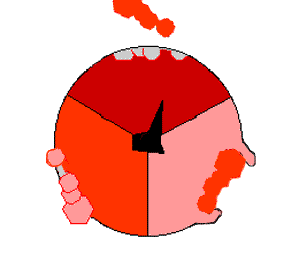
Pumping protons out of the mitochondria creates a proton concentration difference across the membrane and generates an electrochemical gradient. This force drives protons back into the mitochondrion through the base of an enzyme called ATP synthase. The flow of protons makes the stalk subunit rotate, causing the active site of the synthase domain to change shape and phosphorylate adenosine diphosphate – turning it into ATP.
Energy from inorganic compounds
Chemolithotrophy is a type of metabolism found in prokaryotes where energy is obtained from the oxidation of inorganic compounds. These organisms can use hydrogen, reduced sulfur compounds (such as sulfide, hydrogen sulfide and thiosulfate), ferrous iron (Fe(II)) or ammonia as sources of reducing power and they gain energy from the oxidation of these compounds. These microbial processes are important in global biogeochemical cycles such as acetogenesis, nitrification and denitrification and are critical for soil fertility.
Energy from light
The energy in sunlight is captured by plants, cyanobacteria, purple bacteria, green sulfur bacteria and some protists. This process is often coupled to the conversion of carbon dioxide into organic compounds, as part of photosynthesis, which is discussed below. The energy capture and carbon fixation systems can, however, operate separately in prokaryotes, as purple bacteria and green sulfur bacteria can use sunlight as a source of energy, while switching between carbon fixation and the fermentation of organic compounds.
In many organisms, the capture of solar energy is similar in principle to oxidative phosphorylation, as it involves the storage of energy as a proton concentration gradient. This proton motive force then drives ATP synthesis. The electrons needed to drive this electron transport chain come from light-gathering proteins called photosynthetic reaction centres. Reaction centers are classified into two types depending on the nature of photosynthetic pigment present, with most photosynthetic bacteria only having one type, while plants and cyanobacteria have two.
In plants, algae, and cyanobacteria, photosystem II uses light energy to remove electrons from water, releasing oxygen as a waste product. The electrons then flow to the cytochrome b6f complex, which uses their energy to pump protons across the thylakoid membrane in the chloroplast. These protons move back through the membrane as they drive the ATP synthase, as before. The electrons then flow through photosystem I and can then be used to reduce the coenzyme NADP+.
Anabolism
Anabolism is the set of constructive metabolic processes where the energy released by catabolism is used to synthesize complex molecules. In general, the complex molecules that make up cellular structures are constructed step-by-step from smaller and simpler precursors. Anabolism involves three basic stages. First, the production of precursors such as amino acids, monosaccharides, isoprenoids and nucleotides, secondly, their activation into reactive forms using energy from ATP, and thirdly, the assembly of these precursors into complex molecules such as proteins, polysaccharides, lipids and nucleic acids.
Anabolism in organisms can be different according to the source of constructed molecules in their cells. Autotrophs such as plants can construct the complex organic molecules in their cells such as polysaccharides and proteins from simple molecules like carbon dioxide and water. Heterotrophs, on the other hand, require a source of more complex substances, such as monosaccharides and amino acids, to produce these complex molecules. Organisms can be further classified by ultimate source of their energy: photoautotrophs and photoheterotrophs obtain energy from light, whereas chemoautotrophs and chemoheterotrophs obtain energy from oxidation reactions.
Carbon fixation
Photosynthesis is the synthesis of carbohydrates from sunlight and carbon dioxide (CO2). In plants, cyanobacteria and algae, oxygenic photosynthesis splits water, with oxygen produced as a waste product. This process uses the ATP and NADPH produced by the photosynthetic reaction centres, as described above, to convert CO2 into glycerate 3-phosphate, which can then be converted into glucose. This carbon-fixation reaction is carried out by the enzyme RuBisCO as part of the Calvin – Benson cycle. Three types of photosynthesis occur in plants, C3 carbon fixation, C4 carbon fixation and CAM photosynthesis. These differ by the route that carbon dioxide takes to the Calvin cycle, with C3 plants fixing CO2 directly, while C4 and CAM photosynthesis incorporate the CO2 into other compounds first, as adaptations to deal with intense sunlight and dry conditions.
In photosynthetic prokaryotes the mechanisms of carbon fixation are more diverse. Here, carbon dioxide can be fixed by the Calvin – Benson cycle, a reversed citric acid cycle, or the carboxylation of acetyl-CoA. Prokaryotic chemoautotrophs also fix CO2 through the Calvin–Benson cycle, but use energy from inorganic compounds to drive the reaction.
Carbohydrates and glycans
In carbohydrate anabolism, simple organic acids can be converted into monosaccharides such as glucose and then used to assemble polysaccharides such as starch. The generation of glucose from compounds like pyruvate, lactate, glycerol, glycerate 3-phosphate and amino acids is called gluconeogenesis. Gluconeogenesis converts pyruvate to glucose-6-phosphate through a series of intermediates, many of which are shared with glycolysis. However, this pathway is not simply glycolysis run in reverse, as several steps are catalyzed by non-glycolytic enzymes. This is important as it allows the formation and breakdown of glucose to be regulated separately, and prevents both pathways from running simultaneously in a futile cycle.
Although fat is a common way of storing energy, in vertebrates such as humans the fatty acids in these stores cannot be converted to glucose through gluconeogenesis as these organisms cannot convert acetyl-CoA into pyruvate; plants do, but animals do not, have the necessary enzymatic machinery. As a result, after long-term starvation, vertebrates need to produce ketone bodies from fatty acids to replace glucose in tissues such as the brain that cannot metabolize fatty acids. In other organisms such as plants and bacteria, this metabolic problem is solved using the glyoxylate cycle, which bypasses the decarboxylation step in the citric acid cycle and allows the transformation of acetyl-CoA to oxaloacetate, where it can be used for the production of glucose. Other than fat, glucose is stored in most tissues, as an energy resource available within the tissue through glycogenesis which was usually being used to maintained glucose level in blood.
Polysaccharides and glycans are made by the sequential addition of monosaccharides by glycosyltransferase from a reactive sugar-phosphate donor such as uridine diphosphate glucose (UDP-Glc) to an acceptor hydroxyl group on the growing polysaccharide. As any of the hydroxyl groups on the ring of the substrate can be acceptors, the polysaccharides produced can have straight or branched structures. The polysaccharides produced can have structural or metabolic functions themselves, or be transferred to lipids and proteins by enzymes called oligosaccharyltransferases.
Fatty acids, isoprenoids and sterol

Fatty acids are made by fatty acid synthases that polymerize and then reduce acetyl-CoA units. The acyl chains in the fatty acids are extended by a cycle of reactions that add the acyl group, reduce it to an alcohol, dehydrate it to an alkene group and then reduce it again to an alkane group. The enzymes of fatty acid biosynthesis are divided into two groups: in animals and fungi, all these fatty acid synthase reactions are carried out by a single multifunctional type I protein, while in plant plastids and bacteria separate type II enzymes perform each step in the pathway.
Terpenes and isoprenoids are a large class of lipids that include the carotenoids and form the largest class of plant natural products. These compounds are made by the assembly and modification of isoprene units donated from the reactive precursors isopentenyl pyrophosphate and dimethylallyl pyrophosphate. These precursors can be made in different ways. In animals and archaea, the mevalonate pathway produces these compounds from acetyl-CoA, while in plants and bacteria the non-mevalonate pathway uses pyruvate and glyceraldehyde 3-phosphate as substrates. One important reaction that uses these activated isoprene donors is sterol biosynthesis. Here, the isoprene units are joined to make squalene and then folded up and formed into a set of rings to make lanosterol. Lanosterol can then be converted into other sterols such as cholesterol and ergosterol.
Proteins
Organisms vary in their ability to synthesize the 20 common amino acids. Most bacteria and plants can synthesize all twenty, but mammals can only synthesize eleven nonessential amino acids, so nine essential amino acids must be obtained from food. Some simple parasites, such as the bacteria Mycoplasma pneumoniae, lack all amino acid synthesis and take their amino acids directly from their hosts. All amino acids are synthesized from intermediates in glycolysis, the citric acid cycle, or the pentose phosphate pathway. Nitrogen is provided by glutamate and glutamine. Nonessensial amino acid synthesis depends on the formation of the appropriate alpha-keto acid, which is then transaminated to form an amino acid.
Amino acids are made into proteins by being joined in a chain of peptide bonds. Each different protein has a unique sequence of amino acid residues: this is its primary structure. Just as the letters of the alphabet can be combined to form an almost endless variety of words, amino acids can be linked in varying sequences to form a huge variety of proteins. Proteins are made from amino acids that have been activated by attachment to a transfer RNA molecule through an ester bond. This aminoacyl-tRNA precursor is produced in an ATP-dependent reaction carried out by an aminoacyl tRNA synthetase. This aminoacyl-tRNA is then a substrate for the ribosome, which joins the amino acid onto the elongating protein chain, using the sequence information in a messenger RNA.
Nucleotide synthesis and salvage
Nucleotides are made from amino acids, carbon dioxide and formic acid in pathways that require large amounts of metabolic energy. Consequently, most organisms have efficient systems to salvage preformed nucleotides. Purines are synthesized as nucleosides (bases attached to ribose). Both adenine and guanine are made from the precursor nucleoside inosine monophosphate, which is synthesized using atoms from the amino acids glycine, glutamine, and aspartic acid, as well as formate transferred from the coenzyme tetrahydrofolate. Pyrimidines, on the other hand, are synthesized from the base orotate, which is formed from glutamine and aspartate.
Xenobiotics and redox metabolism
All organisms are constantly exposed to compounds that they cannot use as foods and that would be harmful if they accumulated in cells, as they have no metabolic function. These potentially damaging compounds are called xenobiotics. Xenobiotics such as synthetic drugs, natural poisons and antibiotics are detoxified by a set of xenobiotic-metabolizing enzymes. In humans, these include cytochrome P450 oxidases, UDP-glucuronosyltransferases, and glutathione S-transferases. This system of enzymes acts in three stages to firstly oxidize the xenobiotic (phase I) and then conjugate water-soluble groups onto the molecule (phase II). The modified water-soluble xenobiotic can then be pumped out of cells and in multicellular organisms may be further metabolized before being excreted (phase III). In ecology, these reactions are particularly important in microbial biodegradation of pollutants and the bioremediation of contaminated land and oil spills. Many of these microbial reactions are shared with multicellular organisms, but due to the incredible diversity of types of microbes these organisms are able to deal with a far wider range of xenobiotics than multicellular organisms, and can degrade even persistent organic pollutants such as organochloride compounds.
A related problem for aerobic organisms is oxidative stress. Here, processes including oxidative phosphorylation and the formation of disulfide bonds during protein folding produce reactive oxygen species such as hydrogen peroxide. These damaging oxidants are removed by antioxidant metabolites such as glutathione and enzymes such as catalases and peroxidases.
Thermodynamics of living organisms
Living organisms must obey the laws of thermodynamics, which describe the transfer of heat and work. The second law of thermodynamics states that in any isolated system, the amount of entropy (disorder) cannot decrease. Although living organisms' amazing complexity appears to contradict this law, life is possible as all organisms are open systems that exchange matter and energy with their surroundings. Living systems are not in equilibrium, but instead are dissipative systems that maintain their state of high complexity by causing a larger increase in the entropy of their environments. The metabolism of a cell achieves this by coupling the spontaneous processes of catabolism to the non-spontaneous processes of anabolism. In thermodynamic terms, metabolism maintains order by creating disorder.
Regulation and control
As the environments of most organisms are constantly changing, the reactions of metabolism must be finely regulated to maintain a constant set of conditions within cells, a condition called homeostasis. Metabolic regulation also allows organisms to respond to signals and interact actively with their environments. Two closely linked concepts are important for understanding how metabolic pathways are controlled. Firstly, the regulation of an enzyme in a pathway is how its activity is increased and decreased in response to signals. Secondly, the control exerted by this enzyme is the effect that these changes in its activity have on the overall rate of the pathway (the flux through the pathway). For example, an enzyme may show large changes in activity (i.e. it is highly regulated) but if these changes have little effect on the flux of a metabolic pathway, then this enzyme is not involved in the control of the pathway.

There are multiple levels of metabolic regulation. In intrinsic regulation, the metabolic pathway self-regulates to respond to changes in the levels of substrates or products; for example, a decrease in the amount of product can increase the flux through the pathway to compensate. This type of regulation often involves allosteric regulation of the activities of multiple enzymes in the pathway. Extrinsic control involves a cell in a multicellular organism changing its metabolism in response to signals from other cells. These signals are usually in the form of water-soluble messengers such as hormones and growth factors and are detected by specific receptors on the cell surface. These signals are then transmitted inside the cell by second messenger systems that often involved the phosphorylation of proteins.
A very well understood example of extrinsic control is the regulation of glucose metabolism by the hormone insulin. Insulin is produced in response to rises in blood glucose levels. Binding of the hormone to insulin receptors on cells then activates a cascade of protein kinases that cause the cells to take up glucose and convert it into storage molecules such as fatty acids and glycogen. The metabolism of glycogen is controlled by activity of phosphorylase, the enzyme that breaks down glycogen, and glycogen synthase, the enzyme that makes it. These enzymes are regulated in a reciprocal fashion, with phosphorylation inhibiting glycogen synthase, but activating phosphorylase. Insulin causes glycogen synthesis by activating protein phosphatases and producing a decrease in the phosphorylation of these enzymes.
Evolution

The central pathways of metabolism described above, such as glycolysis and the citric acid cycle, are present in all three domains of living things and were present in the last universal common ancestor. This universal ancestral cell was prokaryotic and probably a methanogen that had extensive amino acid, nucleotide, carbohydrate and lipid metabolism. The retention of these ancient pathways during later evolution may be the result of these reactions having been an optimal solution to their particular metabolic problems, with pathways such as glycolysis and the citric acid cycle producing their end products highly efficiently and in a minimal number of steps. The first pathways of enzyme-based metabolism may have been parts of purine nucleotide metabolism, while previous metabolic pathways were a part of the ancient RNA world.
Many models have been proposed to describe the mechanisms by which novel metabolic pathways evolve. These include the sequential addition of novel enzymes to a short ancestral pathway, the duplication and then divergence of entire pathways as well as the recruitment of pre-existing enzymes and their assembly into a novel reaction pathway. The relative importance of these mechanisms is unclear, but genomic studies have shown that enzymes in a pathway are likely to have a shared ancestry, suggesting that many pathways have evolved in a step-by-step fashion with novel functions created from pre-existing steps in the pathway. An alternative model comes from studies that trace the evolution of proteins' structures in metabolic networks, this has suggested that enzymes are pervasively recruited, borrowing enzymes to perform similar functions in different metabolic pathways (evident in the MANET database) A third possibility is that some parts of metabolism might exist as "modules" that can be reused in different pathways and perform similar functions on different molecules.
As well as the evolution of new metabolic pathways, evolution can also cause the loss of metabolic functions. For example, in some parasites metabolic processes that are not essential for survival are lost and preformed amino acids, nucleotides and carbohydrates may instead be scavenged from the host. Similar reduced metabolic capabilities are seen in endosymbiotic organisms.
Investigation and manipulation

Classically, metabolism is studied by a reductionist approach that focuses on a single metabolic pathway. Particularly valuable is the use of radioactive tracers at the whole-organism, tissue and cellular levels, which define the paths from precursors to final products by identifying radioactively labelled intermediates and products. The enzymes that catalyze these chemical reactions can then be purified and their kinetics and responses to inhibitors investigated. A parallel approach is to identify the small molecules in a cell or tissue; the complete set of these molecules is called the metabolome. Overall, these studies give a good view of the structure and function of simple metabolic pathways, but are inadequate when applied to more complex systems such as the metabolism of a complete cell.
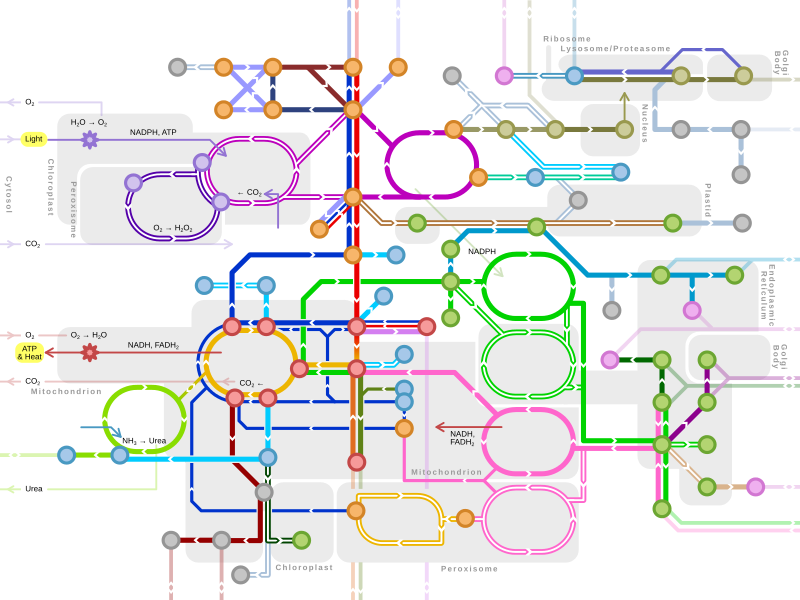
An idea of the complexity of the metabolic networks in cells that contain thousands of different enzymes is given by the figure showing the interactions between just 43 proteins and 40 metabolites to the right: the sequences of genomes provide lists containing anything up to 26.500 genes. However, it is now possible to use this genomic data to reconstruct complete networks of biochemical reactions and produce more holistic mathematical models that may explain and predict their behavior. These models are especially powerful when used to integrate the pathway and metabolite data obtained through classical methods with data on gene expression from proteomic and DNA microarray studies. Using these techniques, a model of human metabolism has now been produced, which will guide future drug discovery and biochemical research. These models are now used in network analysis, to classify human diseases into groups that share common proteins or metabolites.
Bacterial metabolic networks are a striking example of bow-tie organization, an architecture able to input a wide range of nutrients and produce a large variety of products and complex macromolecules using a relatively few intermediate common currencies.
A major technological application of this information is metabolic engineering. Here, organisms such as yeast, plants or bacteria are genetically modified to make them more useful in biotechnology and aid the production of drugs such as antibiotics or industrial chemicals such as 1,3-propanediol and shikimic acid. These genetic modifications usually aim to reduce the amount of energy used to produce the product, increase yields and reduce the production of wastes.
History
The term metabolism is derived from the Ancient Greek word μεταβολή – "Metabole" for "a change" which derived from μεταβάλλ –"Metaballein" means "To change"

Greek philosophy
Aristotle's The Parts of Animals sets out enough details of his views on metabolism for an open flow model to be made. He believed that at each stage of the process, materials from food were transformed, with heat being released as the classical element of fire, and residual materials being excreted as urine, bile, or faeces.
Ibn al-Nafis described metabolism in his 1260 AD work titled Al-Risalah al-Kamiliyyah fil Siera al-Nabawiyyah (The Treatise of Kamil on the Prophet's Biography) which included the following phrase "Both the body and its parts are in a continuous state of dissolution and nourishment, so they are inevitably undergoing permanent change."
Application of the scientific method and Modern metabolic theories
The history of the scientific study of metabolism spans several centuries and has moved from examining whole animals in early studies, to examining individual metabolic reactions in modern biochemistry. The first controlled experiments in human metabolism were published by Santorio Santorio in 1614 in his book Ars de statica medicina. He described how he weighed himself before and after eating, sleep, working, sex, fasting, drinking, and excreting. He found that most of the food he took in was lost through what he called "insensible perspiration".

In these early studies, the mechanisms of these metabolic processes had not been identified and a vital force was thought to animate living tissue. In the 19th century, when studying the fermentation of sugar to alcohol by yeast, Louis Pasteur concluded that fermentation was catalyzed by substances within the yeast cells he called "ferments". He wrote that "alcoholic fermentation is an act correlated with the life and organization of the yeast cells, not with the death or putrefaction of the cells." This discovery, along with the publication by Friedrich Wöhler in 1828 of a paper on the chemical synthesis of urea, and is notable for being the first organic compound prepared from wholly inorganic precursors. This proved that the organic compounds and chemical reactions found in cells were no different in principle than any other part of chemistry.
It was the discovery of enzymes at the beginning of the 20th century by Eduard Buchner that separated the study of the chemical reactions of metabolism from the biological study of cells, and marked the beginnings of biochemistry. The mass of biochemical knowledge grew rapidly throughout the early 20th century. One of the most prolific of these modern biochemists was Hans Krebs who made huge contributions to the study of metabolism. He discovered the urea cycle and later, working with Hans Kornberg, the citric acid cycle and the glyoxylate cycle.
See also
- Anthropogenic metabolism
- Antimetabolite
- Calorimetry
- Isothermal microcalorimetry
- Inborn errors of metabolism
- Iron–sulfur world hypothesis, a "metabolism first" theory of the origin of life
- Metabolic disorder
- Microphysiometry
- Primary nutritional groups
- Proto-metabolism
- Respirometry
- Stream metabolism
- Sulfur metabolism
- Thermic effect of food
- Urban metabolism
- Water metabolism
- Overflow metabolism
- Oncometabolism
- Reactome
- KEGG – Collection of bioinformatics databases
References
Further reading
| Library resources about Metabolism |
Introductory
- Rose S, Mileusnic R (1999). The Chemistry of Life. Penguin Press Science. ISBN 0-14-027273-9.
- Schneider EC, Sagan D (2005). Into the Cool: Energy Flow, Thermodynamics, and Life. University of Chicago Press. ISBN 0-226-73936-8.
- Lane N (2004). Oxygen: The Molecule that Made the World. USA: Oxford University Press. ISBN 0-19-860783-0.
Advanced
- Price N, Stevens L (1999). Fundamentals of Enzymology: Cell and Molecular Biology of Catalytic Proteins. Oxford University Press. ISBN 0-19-850229-X.
- Berg J, Tymoczko J, Stryer L (2002). Biochemistry. W. H. Freeman and Company. ISBN 0-7167-4955-6.
- Cox M, Nelson DL (2004). Lehninger Principles of Biochemistry. Palgrave Macmillan. ISBN 0-7167-4339-6.
- Brock TD, Madigan MR, Martinko J, Parker J (2002). Brock's Biology of Microorganisms. Benjamin Cummings. ISBN 0-13-066271-2.
- Da Silva JJ, Williams RJ (1991). The Biological Chemistry of the Elements: The Inorganic Chemistry of Life. Clarendon Press. ISBN 0-19-855598-9.
- Nicholls DG, Ferguson SJ (2002). Bioenergetics. Academic Press Inc. ISBN 0-12-518121-3.
- Wood HG (February 1991). "Life with CO or CO2 and H2 as a source of carbon and energy". FASEB Journal. 5 (2): 156–63. doi:10.1096/fasebj.5.2.1900793. PMID 1900793. S2CID 45967404.
External links
General information
- The Biochemistry of Metabolism (archived 8 March 2005)
- Sparknotes SAT biochemistry Overview of biochemistry. School level.
- MIT Biology Hypertextbook Undergraduate-level guide to molecular biology.
Human metabolism
- Topics in Medical Biochemistry Guide to human metabolic pathways. School level.
- THE Medical Biochemistry Page Comprehensive resource on human metabolism.
Databases
Metabolic pathways
- Metabolism reference Pathway Archived 23 February 2009 at the Wayback Machine
- The Nitrogen cycle and Nitrogen fixation at the Wayback Machine (archive index)