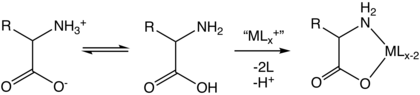
アミノ酸
Amino acid/ja

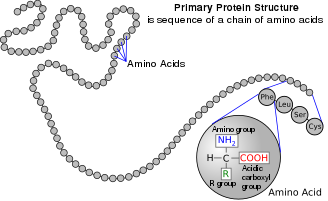
アミノ酸は、アミノ基とカルボン酸基官能基の両方を含む有機化合物である。自然界には500種類以上のアミノ酸が存在するが、タンパク質に組み込まれる22種類のα-アミノ酸が圧倒的に重要である。生命の遺伝コードに登場するのはこの22種類だけである。
アミノ酸は、核となる構造官能基の位置によって分類することができる(α- (α-)、β- (β-)、γ- (γ-)アミノ酸など)。他のカテゴリは、極性(polarity)、イオン化、および側鎖基のタイプ(脂肪族、非環式、芳香族、極性(polar)など。 タンパク質の形態において、アミノ酸残基は人間の筋肉や他の組織の2番目に大きな成分(水が最も大きい)となっている。 タンパク質の残基としての役割を超えて、アミノ酸は神経伝達物質の輸送や生合成などの多くのプロセスに関与している。アミノ酸は地球上の生命とその出現において重要な役割を果たしたと考えられている。
アミノ酸は、IUPAC-IUBMBによって正式に命名されている。生化学命名法に関する合同委員会では、図のような架空の「中性」構造を用いて命名している。例えば、アラニンの系統名はCH
3−CH(NH
2)−COOH式に基づく2-アミノプロパン酸である。欧州委員会はこのアプローチを次のように正当化している:
ここで示した系統名と式は、アミノ基が非プロトン化され、カルボキシル基が解離されていない仮想的な形を指している。この慣例は様々な命名上の問題を避けるために有用であるが、これらの構造がアミノ酸分子のかなりの部分を表していることを意味するものではない。
歴史
最初のアミノ酸は1800年代初頭に発見された。1806年、フランスの化学者Louis-Nicolas VauquelinとPierre Jean Robiquetはアスパラガスから化合物を単離し、その後発見された最初のアミノ酸であるアスパラギンと命名された。シスチンは1810年に発見されたが、その単量体であるシステインは1884年まで発見されなかった。グリシンとロイシンは1820年に発見された。発見された20種類の一般的なアミノ酸の最後は、1935年にウィリアム・カミング・ローズによってスレオニンであり、彼はまた必須アミノ酸を決定し、最適な成長のためのすべてのアミノ酸の1日の最低必要量を確立した。
化学的カテゴリーの単一性は1865年にWurtzによって認識されたが、彼はそれに特別な名前を付けなかった。英語で「アミノ酸」という用語が初めて使われたのは1898年のことで、ドイツ語ではそれ以前にAminosäureという用語が使われていた。タンパク質は酵素消化または酸加水分解後にアミノ酸を生成することが発見された。1902年、Emil FischerとFranz Hofmeisterは、タンパク質は多数のアミノ酸から形成され、それによってあるアミノ酸のアミノ基と別のアミノ酸のカルボキシル基との間に結合が形成され、Fischerが「peptideペプチド」と呼んだ直鎖構造になることを独自に提唱した。
一般的な構造
2-、α-、またはα-アミノ酸は一般的な式H
2NCHRCOOHを持つ。ここでRは"側鎖"として知られる有機置換基である。
何百種類もあるアミノ酸のうち、22種類がタンパク原性(「タンパク質を作る」)である。これらの22種類の化合物が組み合わさって、リボソームによって組み立てられる膨大な数のペプチドやタンパク質ができる。非タンパク質原性アミノ酸あるいは修飾アミノ酸は、翻訳後修飾から、あるいは非リボソームペプチド合成中に生じる。
キラリティ
カルボキシル基の隣の炭素原子はα-炭素と呼ばれる。タンパク質原性アミノ酸では、各アミノ酸に固有のアミンとR基または側鎖を持つ。4つの異なる置換基を持つα-炭素は、グリシンを除くすべてのα-アミノ酸において立体異性である。すべての不斉タンパク質アミノ酸はL配置を持つ。これらは「左巻き」のエナンチオマーであり、α炭素の立体異性体を指す。
いくつかのD-アミノ酸(「右巻き」)は自然界で発見されており、例えばバクテリアのエンベロープの中、ニューロモジュレーター(D-セリン)、いくつかの抗生物質の中に含まれている。まれに、D-アミノ酸残基がタンパク質中に見られ、翻訳後修飾としてL-アミノ酸から変換される。
側鎖
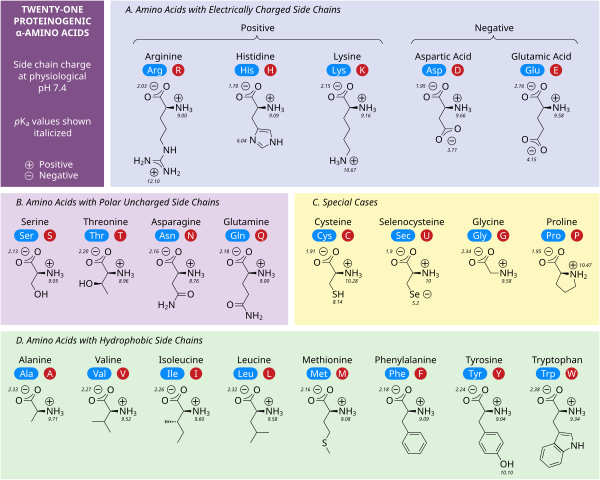
極性荷電側鎖
5つのアミノ酸は中性pHで電荷を持つ。多くの場合、これらの側鎖はタンパク質の表面に現れ、水への溶解を可能にする。反対の電荷を持つ側鎖は、塩橋と呼ばれる重要な静電的接触を形成し、1つのタンパク質内または相互作用するタンパク質間の構造を維持する。多くのタンパク質は金属を特異的にその構造に結合させ、これらの相互作用は一般的にアスパラギン酸、グルタミン酸、ヒスチジンなどの荷電側鎖によって媒介される。特定の条件下では、それぞれのイオン形成基が帯電し、二重塩を形成することがある。
中性pHで負に帯電した2つのアミノ酸は、アスパラギン酸(Asp、D)とグルタミン酸(Glu、E)である。アニオン性カルボキシレート基は、ほとんどの状況でブレンステッド塩基として振る舞う。哺乳類の胃に存在するアスパラギン酸プロテアーゼペプシンのように、非常に低いpH環境にある酵素は、ブレンステッド酸として働くアスパラギン酸残基やグルタミン酸残基を触媒として持つことがある。

アルギニン(Arg、R)、リジン(Lys、K)、ヒスチジン(His、H)の3つである。アルギニンは荷電したグアニジン基を持ち、リジンは荷電したアルキルアミノ基を持ち、pH7で完全にプロトン化される。ヒスチジンのイミダゾール基はpKaが6.0であり、中性pHでは10%程度しかプロトン化されない。ヒスチジンは塩基性と共役酸の形で容易に見出されるため、酵素反応における触媒的プロトン移動にしばしば関与する。
極性荷電側鎖
セリン(Ser, S)、スレオニン(Thr, T)、アスパラギン(Asn, N)、グルタミン(Gln, Q)は、水や他のアミノ酸と容易に水素結合を形成する。通常の状態ではイオン化しないが、セリンプロテアーゼの触媒セリンは例外である。これは深刻な摂動の例であり、一般的なセリン残基の特徴ではない。スレオニンには2つの不斉中心があり、アキラルなグリシンを除くすべてのアミノ酸に共通するα-炭素のL(2S)不斉中心だけでなく、β-炭素の(3R)不斉中心もある。完全な立体化学的仕様は(2S,3R)-L-スレオニンである。
疎水性側鎖
非極性アミノ酸の相互作用は、折りたたみタンパク質が機能的な三次元構造になる過程の主要な原動力である。これらのアミノ酸の側鎖はいずれも容易にイオン化しないため、チロシン(Tyr、Y)を除いてpKaを持たない。チロシンの水酸基は高いpHで脱プロトン化し、負に帯電したフェノラートを形成する。このため、チロシンを極性、非荷電アミノ酸のカテゴリーに入れることもできるが、水への溶解度が非常に低いため、疎水性アミノ酸の特徴とよく一致する。
特別な場合の側鎖
いくつかの側鎖は、荷電性、極性、疎水性のカテゴリーではうまく説明できない。グリシン(Gly, G)は、そのサイズが小さいため、アミノ基とカルボキシレート基によって溶解度が大きく左右されることから、極性アミノ酸と考えられる。しかし、グリシンには側鎖がないため、アミノ酸の中でも独特の柔軟性があり、タンパク質のフォールディングに大きな影響を与える。システイン(Cys, C)も水素結合を形成しやすく、極性アミノ酸のカテゴリーに入るが、タンパク質構造中ではしばしば他のシステインとジスルフィド結合と呼ばれる共有結合を形成している。これらの結合はタンパク質の折り畳みと安定性に影響を与え、抗体の形成に不可欠である。プロリン(Pro, P)はアルキル側鎖を持ち、疎水性であると考えられるが、側鎖がαアミノ基に結合しているため、タンパク質に取り込まれると特に柔軟性がなくなる。グリシンと同様に、これはアミノ酸の中でもユニークな方法でタンパク質の構造に影響を与える。セレノシステイン(Sec, U)はDNAに直接コードされていないが、リボソームを介してタンパク質に組み込まれる珍しいアミノ酸である。セレノシステインは同様のシステインに比べて酸化還元電位が低く、いくつかのユニークな酵素反応に関与している。ピロリジン(Pyl, O)はDNAにコードされていない別のアミノ酸であるが、リボソームによってタンパク質に合成される。古細菌種に存在し、いくつかのメチルトランスフェラーゼの触媒活性に関与している。
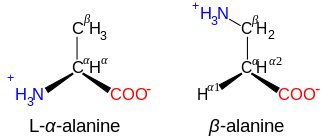
β-アミノ酸とγ-アミノ酸
カルノシンや他のいくつかのペプチドの成分であるβ-アラニンなど、構造NH+
3−CXY−CXY−CO−
2を持つアミノ酸はβ-アミノ酸である。構造NH+
3−CXY−CXY−CXY−CO−
2を持つものはγ-アミノ酸であり、XとYは2つの置換基(一方は通常H)である。
双性イオン

アミノ酸の一般的な天然型は、−NH+
3(プロリンの場合は−NH+
2−)と−CO−
2官能基が同じC原子に結合した双性イオン構造を持つ。リボソームで翻訳される過程でタンパク質中に発見される唯一のものである。
中性に近いpHの水溶液中では、アミノ酸は双性イオン、すなわちNH+
3とCO−
2の両方が荷電状態にある双極子イオンとして存在するため、全体の構造はNH+
3−CHR−CO−
2.となる。生理的pHでは、いわゆる「中性型」−NH
2−CHR−CO
2Hは測定可能な程度では存在しない。双性イオン構造における2つの電荷はゼロに加算されるが、正味電荷がゼロの種を「荷電していない」と呼ぶのは誤解を招く。
強酸性条件下(pH3以下)では、カルボン酸基がプロトン化され、構造はアンモニオカルボン酸NH+
3−CHR−CO
2Hとなる。これは、哺乳類の胃やリソソームなどの酸性環境で活性を発揮するペプシンのような酵素には関係するが、細胞内酵素にはあまり当てはまらない。高塩基性条件下(pH10以上、生理的条件下では通常見られない)では、アンモニオ基は脱プロトン化されてNH
2−CHR−CO−
2を与える。
酸や塩基の定義は、化学の世界では様々なものが使われているが、水溶液中の化学に有用なのは、ブレンステッドの理論だけである:酸とは、他の種にプロトンを供与できる種であり、塩基とは、プロトンを受容できる種である。この基準を用いて、上の図では基を表示している。アスパラギン酸残基とグルタミン酸残基のカルボン酸側鎖は、タンパク質中の主要なブレンステッド塩基である。同様に、リジン、チロシン、システインは通常ブレンステッド酸として働く。これらの条件下では、ヒスチジンはブレンステッド酸としても塩基としても働く。
等電点

For amino acids with uncharged side-chains the zwitterion predominates at pH values between the two pKa values, but coexists in equilibrium with small amounts of net negative and net positive ions. At the midpoint between the two pKa values, the trace amount of net negative and trace of net positive ions balance, so that average net charge of all forms present is zero. This pH is known as the isoelectric point pI, so pI = 1/2(pKa1 + pKa2).
For amino acids with charged side chains, the pKa of the side chain is involved. Thus for aspartate or glutamate with negative side chains, the terminal amino group is essentially entirely in the charged form −NH+
3, but this positive charge needs to be balanced by the state with just one C-terminal carboxylate group is negatively charged. This occurs halfway between the two carboxylate pKa values: pI = 1/2(pKa1 + pKa(R)), where pKa(R) is the side chain pKa.
Similar considerations apply to other amino acids with ionizable side-chains, including not only glutamate (similar to aspartate), but also cysteine, histidine, lysine, tyrosine and arginine with positive side chains.
Amino acids have zero mobility in electrophoresis at their isoelectric point, although this behaviour is more usually exploited for peptides and proteins than single amino acids. Zwitterions have minimum solubility at their isoelectric point, and some amino acids (in particular, with nonpolar side chains) can be isolated by precipitation from water by adjusting the pH to the required isoelectric point.
Physicochemical properties
The 20 canonical amino acids can be classified according to their properties. Important factors are charge, hydrophilicity or hydrophobicity, size, and functional groups. These properties influence protein structure and protein–protein interactions. The water-soluble proteins tend to have their hydrophobic residues (Leu, Ile, Val, Phe, and Trp) buried in the middle of the protein, whereas hydrophilic side chains are exposed to the aqueous solvent. (In biochemistry, a residue refers to a specific monomer within the polymeric chain of a polysaccharide, protein or nucleic acid.) The integral membrane proteins tend to have outer rings of exposed hydrophobic amino acids that anchor them in the lipid bilayer. Some peripheral membrane proteins have a patch of hydrophobic amino acids on their surface that sticks to the membrane. In a similar fashion, proteins that have to bind to positively charged molecules have surfaces rich in negatively charged amino acids such as glutamate and aspartate, while proteins binding to negatively charged molecules have surfaces rich in positively charged amino acids like lysine and arginine. For example, lysine and arginine are present in large amounts in the low-complexity regions of nucleic-acid binding proteins. There are various hydrophobicity scales of amino acid residues.
Some amino acids have special properties. Cysteine can form covalent disulfide bonds to other cysteine residues. Proline forms a cycle to the polypeptide backbone, and glycine is more flexible than other amino acids.
Glycine and proline are strongly present within low complexity regions of both eukaryotic and prokaryotic proteins, whereas the opposite is the case with cysteine, phenylalanine, tryptophan, methionine, valine, leucine, isoleucine, which are highly reactive, or complex, or hydrophobic.
Many proteins undergo a range of posttranslational modifications, whereby additional chemical groups are attached to the amino acid residue side chains sometimes producing lipoproteins (that are hydrophobic), or glycoproteins (that are hydrophilic) allowing the protein to attach temporarily to a membrane. For example, a signaling protein can attach and then detach from a cell membrane, because it contains cysteine residues that can have the fatty acid palmitic acid added to them and subsequently removed.
Table of standard amino acid abbreviations and properties
Although one-letter symbols are included in the table, IUPAC–IUBMB recommend that "Use of the one-letter symbols should be restricted to the comparison of long sequences".
The one-letter notation was chosen by IUPAC-IUB based on the following rules:
- Initial letters are used where there is no ambuiguity: C cysteine, H histidine, I isoleucine, M methionine, S serine, V valine,
- Where arbitrary assignment is needed, the structurally simpler amino acids are given precedence: A Alanine, G glycine, L leucine, P proline, T threonine,
- F PHenylalanine and R aRginine are assigned by being phonetically suggestive,
- W tryptophane is assigned based on the double ring being visually suggestive to the bulky letter W,
- K lysine and Y tyrosine are assigned as alphabetically nearest to their initials L and T (note that U was avoided for its similarity with V, while X was reserved for undetermined or atypical amino acids); for tyrosine the mnemonic tYrosine was also proposed,
- D aspartate was assigned arbitrarily, with the proposed mnemonic asparDic acid; E glutamate was assigned in alphabetical sequence being larger by merely one methylene –CH2– group,
- N asparagine was assigned arbitrarily, with the proposed mnemonic asparagiNe; Q glutamine was assigned in alphabetical sequence of those still available (note again that O was avoided due to similarity with D), with the proposed mnemonic Qlutamine.
| Amino acid | 3- and 1-letter symbols | Side chain | Hydropathy index |
Molar absorptivity | Molecular mass | Abundance in proteins (%) |
Standard genetic coding, IUPAC notation | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | 1 | Class | Chemical polarity | Net charge at pH 7.4 |
Wavelength, λmax (nm) |
Coefficient ε (mM−1·cm−1) | |||||
| Alanine | Ala | A | Aliphatic | Nonpolar | Neutral | 1.8 | 89.094 | 8.76 | GCN | ||
| Arginine | Arg | R | Fixed cation | Basic polar | Positive | −4.5 | 174.203 | 5.78 | MGR, CGY | ||
| Asparagine | Asn | N | Amide | Polar | Neutral | −3.5 | 132.119 | 3.93 | AAY | ||
| Aspartate | Asp | D | Anion | Brønsted base | Negative | −3.5 | 133.104 | 5.49 | GAY | ||
| Cysteine | Cys | C | Thiol | Brønsted acid | Neutral | 2.5 | 250 | 0.3 | 121.154 | 1.38 | UGY |
| Glutamine | Gln | Q | Amide | Polar | Neutral | −3.5 | 146.146 | 3.9 | CAR | ||
| Glutamate | Glu | E | Anion | Brønsted base | Negative | −3.5 | 147.131 | 6.32 | GAR | ||
| Glycine | Gly | G | Aliphatic | Nonpolar | Neutral | −0.4 | 75.067 | 7.03 | GGN | ||
| Histidine | His | H | Cationic | Brønsted acid and base | Positive, 10% Neutral, 90% |
−3.2 | 211 | 5.9 | 155.156 | 2.26 | CAY |
| Isoleucine | Ile | I | Aliphatic | Nonpolar | Neutral | 4.5 | 131.175 | 5.49 | AUH | ||
| Leucine | Leu | L | Aliphatic | Nonpolar | Neutral | 3.8 | 131.175 | 9.68 | YUR, CUY | ||
| Lysine | Lys | K | Cation | Brønsted acid | Positive | −3.9 | 146.189 | 5.19 | AAR | ||
| Methionine | Met | M | Thioether | Nonpolar | Neutral | 1.9 | 149.208 | 2.32 | AUG | ||
| Phenylalanine | Phe | F | Aromatic | Nonpolar | Neutral | 2.8 | 257, 206, 188 | 0.2, 9.3, 60.0 | 165.192 | 3.87 | UUY |
| Proline | Pro | P | Cyclic | Nonpolar | Neutral | −1.6 | 115.132 | 5.02 | CCN | ||
| Serine | Ser | S | Hydroxylic | Polar | Neutral | −0.8 | 105.093 | 7.14 | UCN, AGY | ||
| Threonine | Thr | T | Hydroxylic | Polar | Neutral | −0.7 | 119.119 | 5.53 | ACN | ||
| Tryptophan | Trp | W | Aromatic | Nonpolar | Neutral | −0.9 | 280, 219 | 5.6, 47.0 | 204.228 | 1.25 | UGG |
| Tyrosine | Tyr | Y | Aromatic | Brønsted acid | Neutral | −1.3 | 274, 222, 193 | 1.4, 8.0, 48.0 | 181.191 | 2.91 | UAY |
| Valine | Val | V | Aliphatic | Nonpolar | Neutral | 4.2 | 117.148 | 6.73 | GUN | ||
Two additional amino acids are in some species coded for by codons that are usually interpreted as stop codons:
| 21st and 22nd amino acids | 3-letter | 1-letter | Molecular mass |
|---|---|---|---|
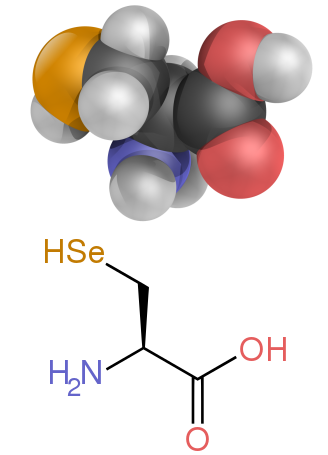
| Selenocysteine | Sec | U | 168.064 |
| Pyrrolysine | Pyl | O | 255.313 |
In addition to the specific amino acid codes, placeholders are used in cases where chemical or crystallographic analysis of a peptide or protein cannot conclusively determine the identity of a residue. They are also used to summarize conserved protein sequence motifs. The use of single letters to indicate sets of similar residues is similar to the use of abbreviation codes for degenerate bases.
| Ambiguous amino acids | 3-letter | 1-letter | Amino acids included | Codons included |
|---|---|---|---|---|
| Any / unknown | Xaa | X | All | NNN |
| Asparagine or aspartate | Asx | B | D, N | RAY |
| Glutamine or glutamate | Glx | Z | E, Q | SAR |
| Leucine or isoleucine | Xle | J | I, L | YTR, ATH, CTY |
| Hydrophobic | Φ | V, I, L, F, W, Y, M | NTN, TAY, TGG | |
| Aromatic | Ω | F, W, Y, H | YWY, TTY, TGG | |
| Aliphatic (non-aromatic) | Ψ | V, I, L, M | VTN, TTR | |
| Small | π | P, G, A, S | BCN, RGY, GGR | |
| Hydrophilic | ζ | S, T, H, N, Q, E, D, K, R | VAN, WCN, CGN, AGY | |
| Positively-charged | + | K, R, H | ARR, CRY, CGR | |
| Negatively-charged | − | D, E | GAN |
Unk is sometimes used instead of Xaa, but is less standard.
Ter or * (from termination) is used in notation for mutations in proteins when a stop codon occurs. It corresponds to no amino acid at all.
In addition, many nonstandard amino acids have a specific code. For example, several peptide drugs, such as Bortezomib and MG132, are artificially synthesized and retain their protecting groups, which have specific codes. Bortezomib is Pyz–Phe–boroLeu, and MG132 is Z–Leu–Leu–Leu–al. To aid in the analysis of protein structure, photo-reactive amino acid analogs are available. These include photoleucine (pLeu) and photomethionine (pMet).
Occurrence and functions in biochemistry
Proteinogenic amino acids
Amino acids are the precursors to proteins. They join by condensation reactions to form short polymer chains called peptides or longer chains called either polypeptides or proteins. These chains are linear and unbranched, with each amino acid residue within the chain attached to two neighboring amino acids. In nature, the process of making proteins encoded by DNA/RNA genetic material is called translation and involves the step-by-step addition of amino acids to a growing protein chain by a ribozyme that is called a ribosome The order in which the amino acids are added is read through the genetic code from an mRNA template, which is an RNA copy of one of the organism's genes.
Twenty-two amino acids are naturally incorporated into polypeptides and are called proteinogenic or natural amino acids. Of these, 20 are encoded by the universal genetic code. The remaining 2, selenocysteine and pyrrolysine, are incorporated into proteins by unique synthetic mechanisms. Selenocysteine is incorporated when the mRNA being translated includes a SECIS element, which causes the UGA codon to encode selenocysteine instead of a stop codon. Pyrrolysine is used by some methanogenic archaea in enzymes that they use to produce methane. It is coded for with the codon UAG, which is normally a stop codon in other organisms. This UAG codon is followed by a PYLIS downstream sequence.
Several independent evolutionary studies have suggested that Gly, Ala, Asp, Val, Ser, Pro, Glu, Leu, Thr may belong to a group of amino acids that constituted the early genetic code, whereas Cys, Met, Tyr, Trp, His, Phe may belong to a group of amino acids that constituted later additions of the genetic code.
Standard vs nonstandard amino acids
The 20 amino acids that are encoded directly by the codons of the universal genetic code are called standard or canonical amino acids. A modified form of methionine (N-formylmethionine) is often incorporated in place of methionine as the initial amino acid of proteins in bacteria, mitochondria and chloroplasts. Other amino acids are called nonstandard or non-canonical. Most of the nonstandard amino acids are also non-proteinogenic (i.e. they cannot be incorporated into proteins during translation), but two of them are proteinogenic, as they can be incorporated translationally into proteins by exploiting information not encoded in the universal genetic code.
The two nonstandard proteinogenic amino acids are selenocysteine (present in many non-eukaryotes as well as most eukaryotes, but not coded directly by DNA) and pyrrolysine (found only in some archaea and at least one bacterium). The incorporation of these nonstandard amino acids is rare. For example, 25 human proteins include selenocysteine in their primary structure, and the structurally characterized enzymes (selenoenzymes) employ selenocysteine as the catalytic moiety in their active sites. Pyrrolysine and selenocysteine are encoded via variant codons. For example, selenocysteine is encoded by stop codon and SECIS element.
N-formylmethionine (which is often the initial amino acid of proteins in bacteria, mitochondria, and chloroplasts) is generally considered as a form of methionine rather than as a separate proteinogenic amino acid. Codon–tRNA combinations not found in nature can also be used to "expand" the genetic code and form novel proteins known as alloproteins incorporating non-proteinogenic amino acids.
Non-proteinogenic amino acids
Aside from the 22 proteinogenic amino acids, many non-proteinogenic amino acids are known. Those either are not found in proteins (for example carnitine, GABA, levothyroxine) or are not produced directly and in isolation by standard cellular machinery. For example, hydroxyproline , is synthesised from proline. Another example is selenomethionine).
Non-proteinogenic amino acids that are found in proteins are formed by post-translational modification. Such modifications can also determine the localization of the protein, e.g., the addition of long hydrophobic groups can cause a protein to bind to a phospholipid membrane. Examples:
- the carboxylation of glutamate allows for better binding of calcium cations,
- Hydroxyproline, generated by hydroxylation of proline, is a major component of the connective tissue collagen.
- Hypusine in the translation initiation factor EIF5A, contains a modification of lysine.
Some non-proteinogenic amino acids are not found in proteins. Examples include 2-aminoisobutyric acid and the neurotransmitter gamma-aminobutyric acid. Non-proteinogenic amino acids often occur as intermediates in the metabolic pathways for standard amino acids – for example, ornithine and citrulline occur in the urea cycle, part of amino acid catabolism (see below). A rare exception to the dominance of α-amino acids in biology is the β-amino acid beta alanine (3-aminopropanoic acid), which is used in plants and microorganisms in the synthesis of pantothenic acid (vitamin B5), a component of coenzyme A.
In mammalian nutrition
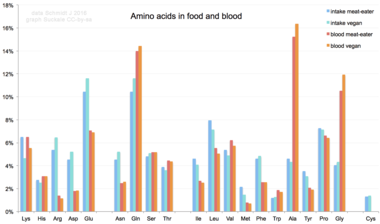
Amino acids are not typical component of food: animals eat proteins. The protein is broken down into amino acids in the process of digestion. They are then used to synthesize new proteins, other biomolecules, or are oxidized to urea and carbon dioxide as a source of energy. The oxidation pathway starts with the removal of the amino group by a transaminase; the amino group is then fed into the urea cycle. The other product of transamidation is a keto acid that enters the citric acid cycle. Glucogenic amino acids can also be converted into glucose, through gluconeogenesis.
Of the 20 standard amino acids, nine (His, Ile, Leu, Lys, Met, Phe, Thr, Trp and Val) are called essential amino acids because the human body cannot synthesize them from other compounds at the level needed for normal growth, so they must be obtained from food.
Semi-essential and conditionally essential amino acids, and juvenile requirements
In addition, cysteine, tyrosine, and arginine are considered semiessential amino acids, and taurine a semi-essential aminosulfonic acid in children. Some amino acids are conditionally essential for certain ages or medical conditions. Essential amino acids may also vary from species to species. The metabolic pathways that synthesize these monomers are not fully developed.
Non-protein functions
Many proteinogenic and non-proteinogenic amino acids have biological functions beyond being precursors to proteins and peptides.In humans, amino acids also have important roles in diverse biosynthetic pathways. Defenses against herbivores in plants sometimes employ amino acids. Examples:
Standard amino acids
- Tryptophan is a precursor of the neurotransmitter serotonin.
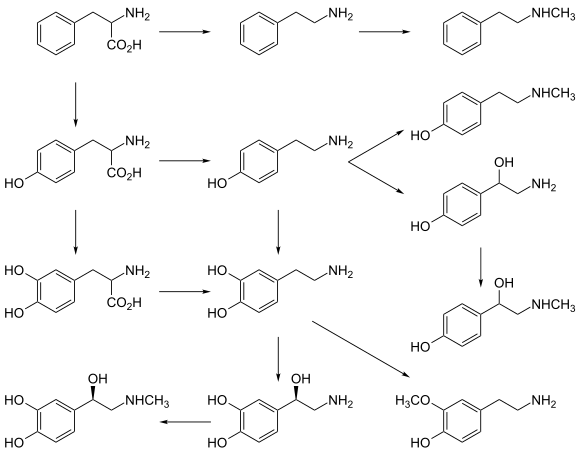
- Tyrosine (and its precursor phenylalanine) are precursors of the catecholamine neurotransmitters dopamine, epinephrine and norepinephrine and various trace amines.
- Phenylalanine is a precursor of phenethylamine and tyrosine in humans. In plants, it is a precursor of various phenylpropanoids, which are important in plant metabolism.
- Glycine is a precursor of porphyrins such as heme.
- Arginine is a precursor of nitric oxide.
- Ornithine and S-adenosylmethionine are precursors of polyamines.
- Aspartate, glycine, and glutamine are precursors of nucleotides. However, not all of the functions of other abundant nonstandard amino acids are known.
Roles for nonstandard amino acids
- Carnitine is used in lipid transport.
- gamma-aminobutyric acid is a neurotransmitter.
- 5-HTP (5-hydroxytryptophan) is used for experimental treatment of depression.
- L-DOPA (L-dihydroxyphenylalanine) for Parkinson's treatment,
- Eflornithine inhibits ornithine decarboxylase and used in the treatment of sleeping sickness.
- Canavanine, an analogue of arginine found in many legumes is an antifeedant, protecting the plant from predators.
- Mimosine found in some legumes, is another possible antifeedant. This compound is an analogue of tyrosine and can poison animals that graze on these plants.
Uses in industry
Animal feed
Amino acids are sometimes added to animal feed because some of the components of these feeds, such as soybeans, have low levels of some of the essential amino acids, especially of lysine, methionine, threonine, and tryptophan. Likewise amino acids are used to chelate metal cations in order to improve the absorption of minerals from feed supplements.
Food
The food industry is a major consumer of amino acids, especially glutamic acid, which is used as a flavor enhancer, and aspartame (aspartylphenylalanine 1-methyl ester), which is used as an artificial sweetener. Amino acids are sometimes added to food by manufacturers to alleviate symptoms of mineral deficiencies, such as anemia, by improving mineral absorption and reducing negative side effects from inorganic mineral supplementation.
Chemical building blocks
Amino acids are low-cost feedstocks used in chiral pool synthesis as enantiomerically pure building blocks.
Amino acids are used in the synthesis of some cosmetics.
Aspirational uses
Fertilizer
The chelating ability of amino acids is sometimes used in fertilizers to facilitate the delivery of minerals to plants in order to correct mineral deficiencies, such as iron chlorosis. These fertilizers are also used to prevent deficiencies from occurring and to improve the overall health of the plants.
Biodegradable plastics
Amino acids have been considered as components of biodegradable polymers, which have applications as environmentally friendly packaging and in medicine in drug delivery and the construction of prosthetic implants. An interesting example of such materials is polyaspartate, a water-soluble biodegradable polymer that may have applications in disposable diapers and agriculture. Due to its solubility and ability to chelate metal ions, polyaspartate is also being used as a biodegradable antiscaling agent and a corrosion inhibitor.
Chemical synthesis
The commercial production of amino acids usually relies on mutant bacteria that overproduce individual amino acids using glucose as a carbon source. Some amino acids are produced by enzymatic conversions of synthetic intermediates. 2-Aminothiazoline-4-carboxylic acid is an intermediate in one industrial synthesis of L-cysteine for example. Aspartic acid is produced by the addition of ammonia to fumarate using a lyase.
Biosynthesis
In plants, nitrogen is first assimilated into organic compounds in the form of glutamate, formed from alpha-ketoglutarate and ammonia in the mitochondrion. For other amino acids, plants use transaminases to move the amino group from glutamate to another alpha-keto acid. For example, aspartate aminotransferase converts glutamate and oxaloacetate to alpha-ketoglutarate and aspartate. Other organisms use transaminases for amino acid synthesis, too.
Nonstandard amino acids are usually formed through modifications to standard amino acids. For example, homocysteine is formed through the transsulfuration pathway or by the demethylation of methionine via the intermediate metabolite S-adenosylmethionine, while hydroxyproline is made by a post translational modification of proline.
Microorganisms and plants synthesize many uncommon amino acids. For example, some microbes make 2-aminoisobutyric acid and lanthionine, which is a sulfide-bridged derivative of alanine. Both of these amino acids are found in peptidic lantibiotics such as alamethicin. However, in plants, 1-aminocyclopropane-1-carboxylic acid is a small disubstituted cyclic amino acid that is an intermediate in the production of the plant hormone ethylene.
Primordial synthesis
The formation of amino acids and peptides are assumed to precede and perhaps induce the emergence of life on earth. Amino acids can form from simple precursors under various conditions. Surface-based chemical metabolism of amino acids and very small compounds may have led to the build-up of amino acids, coenzymes and phosphate-based small carbon molecules. Amino acids and similar building blocks could have been elaborated into proto-peptides, with peptides being considered key players in the origin of life.
In the famous Urey-Miller experiment, the passage of an electric arc through a mixture of methane, hydrogen, and ammonia produces a large number of amino acids. Since then, scientists have discovered a range of ways and components by which the potentially prebiotic formation and chemical evolution of peptides may have occurred, such as condensing agents, the design of self-replicating peptides and a number of non-enzymatic mechanisms by which amino acids could have emerged and elaborated into peptides. Several hypotheses invoke the Strecker synthesis whereby hydrogen cyanide, simple aldehydes, ammonia, and water produce amino acids.
According to a review, amino acids, and even peptides, "turn up fairly regularly in the various experimental broths that have been allowed to be cooked from simple chemicals. This is because nucleotides are far more difficult to synthesize chemically than amino acids." For a chronological order, it suggests that there must have been a 'protein world' or at least a 'polypeptide world', possibly later followed by the 'RNA world' and the 'DNA world'. Codon–amino acids mappings may be the biological information system at the primordial origin of life on Earth. While amino acids and consequently simple peptides must have formed under different experimentally probed geochemical scenarios, the transition from an abiotic world to the first life forms is to a large extent still unresolved.
Reactions
Amino acids undergo the reactions expected of the constituent functional groups.
Peptide bond formation
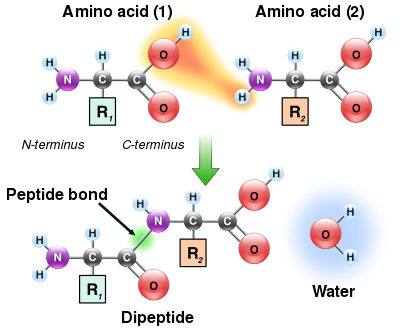
As both the amine and carboxylic acid groups of amino acids can react to form amide bonds, one amino acid molecule can react with another and become joined through an amide linkage. This polymerization of amino acids is what creates proteins. This condensation reaction yields the newly formed peptide bond and a molecule of water. In cells, this reaction does not occur directly; instead, the amino acid is first activated by attachment to a transfer RNA molecule through an ester bond. This aminoacyl-tRNA is produced in an ATP-dependent reaction carried out by an aminoacyl tRNA synthetase. This aminoacyl-tRNA is then a substrate for the ribosome, which catalyzes the attack of the amino group of the elongating protein chain on the ester bond. As a result of this mechanism, all proteins made by ribosomes are synthesized starting at their N-terminus and moving toward their C-terminus.
However, not all peptide bonds are formed in this way. In a few cases, peptides are synthesized by specific enzymes. For example, the tripeptide glutathione is an essential part of the defenses of cells against oxidative stress. This peptide is synthesized in two steps from free amino acids. In the first step, gamma-glutamylcysteine synthetase condenses cysteine and glutamate through a peptide bond formed between the side chain carboxyl of the glutamate (the gamma carbon of this side chain) and the amino group of the cysteine. This dipeptide is then condensed with glycine by glutathione synthetase to form glutathione.
In chemistry, peptides are synthesized by a variety of reactions. One of the most-used in solid-phase peptide synthesis uses the aromatic oxime derivatives of amino acids as activated units. These are added in sequence onto the growing peptide chain, which is attached to a solid resin support. Libraries of peptides are used in drug discovery through high-throughput screening.
The combination of functional groups allow amino acids to be effective polydentate ligands for metal–amino acid chelates. The multiple side chains of amino acids can also undergo chemical reactions.
Catabolism
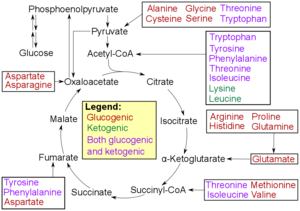
* Glucogenic, with the products having the ability to form glucose by gluconeogenesis
* Ketogenic, with the products not having the ability to form glucose. These products may still be used for ketogenesis or lipid synthesis.
* Amino acids catabolized into both glucogenic and ketogenic products.
Degradation of an amino acid often involves deamination by moving its amino group to α-ketoglutarate, forming glutamate. This process involves transaminases, often the same as those used in amination during synthesis. In many vertebrates, the amino group is then removed through the urea cycle and is excreted in the form of urea. However, amino acid degradation can produce uric acid or ammonia instead. For example, serine dehydratase converts serine to pyruvate and ammonia. After removal of one or more amino groups, the remainder of the molecule can sometimes be used to synthesize new amino acids, or it can be used for energy by entering glycolysis or the citric acid cycle, as detailed in image at right.
Complexation
Amino acids are bidentate ligands, forming transition metal amino acid complexes.
化学分析
有機物に含まれる全窒素は、主にタンパク質中のアミノ基によって形成される。全ケルダール窒素(TKN)は、(排水)、土壌、食品、飼料、有機物全般の分析に広く用いられる窒素の指標である。その名の通り、ケルダール法が適用される。より感度の高い方法もある。
こちらも参照
さらに読む
- Tymoczko JL (2012). "Protein Composition and Structure". Biochemistry. New York: W. H. Freeman and company. pp. 28–31. ISBN 9781429229364.
- Doolittle RF (1989). "Redundancies in protein sequences". In Fasman GD (ed.). Predictions of Protein Structure and the Principles of Protein Conformation. New York: Plenum Press. pp. 599–623. ISBN 978-0-306-43131-9. LCCN 89008555.
- Nelson DL, Cox MM (2000). Lehninger Principles of Biochemistry (3rd ed.). Worth Publishers. ISBN 978-1-57259-153-0. LCCN 99049137.
- Meierhenrich U (2008). Amino acids and the asymmetry of life (PDF). Berlin: Springer Verlag. ISBN 978-3-540-76885-2. LCCN 2008930865. Archived from the original (PDF) on 12 January 2012.
外部リンク
 Media related to Amino acid at Wikimedia Commons
Media related to Amino acid at Wikimedia Commons